Gsas Rod
A Medium Sized Organic Molecule
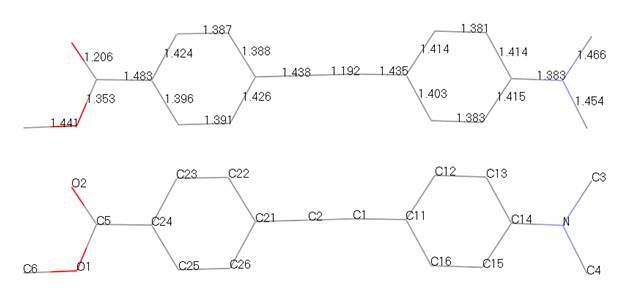
Aim of tutorial: this problem shows the refinement of an organic molecule. The data were collected in a couple of hours on a laboratory diffractometer. The data files are d8_00796.gsas, and rod.inst. Coordinates and atomic labeling from a simulated annealing run to solve the structure are shown below and contained in rod_noh.cif. The atom sequence numbers (for planar restraints) are also listed.
Instructions
1. Read in the molecule from rod_noh.cif (21 atoms) and refine cell parameters, zero point, profile, and overall temperature factor and scale factor. You should get chi**2 below 6 with 9 background terms.
2. Set up some sensible restraints. e.g make each of the rings planar with equal bond lengths, restrain all other bond distances to sensible values. Start with weight factors on restraints of 20.
3. Refine with atom damping of 9. Slowly reduce the weighting factor on restraints. You should be able to get chi**2 below 4.
4. Output a .cif file and look at the molecule.
Outcome
This example should show some of the difficulties of refining organic molecules! Compare your refined structure with rod_singlecrystal.cif which contains single crystal coordinates.
Additional Work (optional)
Try playing with weighting factors to see how they influence R-factors and the shape of the molecule. If you’re familiar with single crystal packages try introducing H atoms and let them ride on C atoms (they do have a significant influence on low angle reflections in this example). Try importing rod.cif and doing refinements with/without hydrogen atoms to see how much they influence a powder pattern.
Try the topas tutorial on the same example.
